ProfAff
Development of an Affinity Interactomic Database
Scientific Context
Molecular complexes play a central role in cellular processes and can be studied using targeted approaches, enabling a deep understanding of specific molecular systems. However, interactomics, which investigates the entirety of molecular interaction networks, remains largely qualitative. These networks are often represented as graphs without precise quantification of the affinities between molecular partners. This lack of quantitative data limits our ability to fully understand cellular dynamics and biological mechanisms.To address this limitation, the Quantitative Interactomics and Disease-Related Networks team, established in 2024 at iBV, is developing innovative systemic biochemistry approaches. These methods allow for large-scale mapping of protein interactions and their molecular affinities. The research team’s work has already produced an extensive dataset of over 5 million protein affinities, enabling the modeling of differences in interaction networks across various cellular contexts and predicting the self-organizing properties of these networks. The team’s ambitious goal is to reach 500 million affinity measurements in the coming years.
To centralize this wealth of data, a groundbreaking database called ProfAff (Profiling Affinities) has been developed.
Project Objectives
The Development of an Affinity Interactomic Database project aims to design innovative experimental and analytical methods for studying protein interactions. By combining the strengths of quantitative biochemistry and interactomics, the project seeks to create a comprehensive database cataloging protein interactions and their affinities.Key milestones of this project, in partnership with the MSI, include:
- Developing a computational workflow to automate data preprocessing;
- Adapting the ProfAff database for the storage and analysis of large-scale datasets;
- Deploying the database, hosted by Université Côte d’Azur.
Expected Impact
The project will deliver a public, quantitative, and comprehensive database that will revolutionize the exploration of protein interaction networks. This resource will simplify the study of biological and pathological processes while opening new avenues for biomedical research and therapeutic development.
Project Leader
Gergo Gogl, Quantitative Interactomics and Disease-Related Networks Team Leader at iBV – Institut de Biologie de Valrose, Université Côte d’Azur.
Project Members
Milissa Abboute, Intern. Master’s student at Université Côte d’Azur.Carole Belliardo, Research Engineer at the MSI – Center of Modeling, Simulation and Interactions, Université Côte d’Azur.
Boglarka Zambo, Postdoctoral Fellow at iBV – Institut de Biologie de Valrose, Université Côte d’Azur.
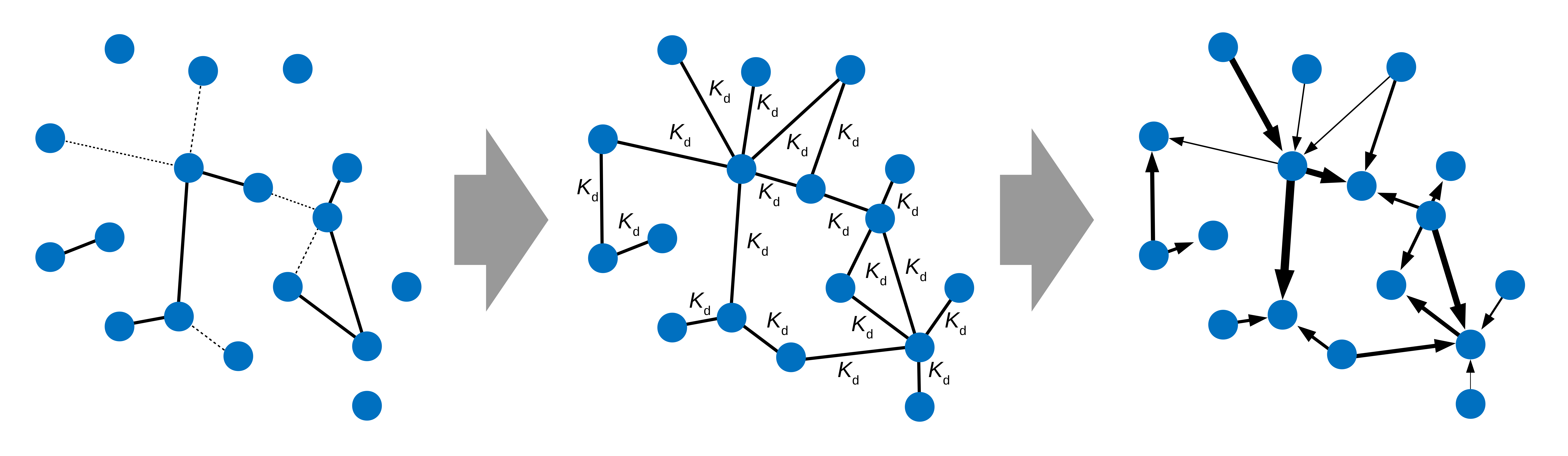
Interaction networks are typically studied using qualitative tools that only allow the capture of the most stable interactions.
System biochemistry allows to study complete networks and survey binding parameters across them. These biochemical properties allow us to study complex networks from a functional aspect.

