MetaBinMG
Enhancing Soil Metagenome reconstruction using combined coverage information for binning
The MetaBinMG project is part of the research work carried out by the Priority Research Projects Microbiomes of Cultivated Plants and Digital Technologies (PEPR MISTIC) consortium, which aims to develop tools for studying the microbial diversity associated with plant crops and their health.
Preliminary work reveals that current analysis strategies fail to adequately represent the genomes of soil microbial communities. Indeed, the various sequencing technologies available seem to capture only a fraction of the true diversity of these complex environmental samples. In addition, bioinformatics methods offer only partial reconstruction of microbial genomes.
This mission has two main objectives. First, it seeks to improve genome reconstruction by developing innovative strategies to fully exploit the information contained in the various available metagenomic data. Second, it plans to use currently produced metabarcoding data to assess the proportion of diversity captured by metagenomic data.
These results will precisely size the metagenomic sequencing efforts required to reconstruct a set of genomes representative of the community of microorganisms in environmental samples.
Project Leader:
Etienne Danchin, Institut Sophia Agrobiotech
Project Partners:
-
Institut Sophia Agrobiotech (Etienne Danchin, Marc Bailly-Bechet)
-
MSI – Center of Modeling, Simulation and Interactions (Carole Belliardo)
-
Inria Center at the University of Bordeaux (David Sherman, Clémence Frioux, and Alain Franc - Pleiade Project Team; Erwan Grichoux, PGTB Platform Manager)
-
Inria Center at Rennes University (Nicolas Maurice, Claire Lemaitre, Riccardo Vicedomini)
-
Burgundy-Franche-Comté INRAE Center (Samuel Mondy, Genosol Platform Manager)
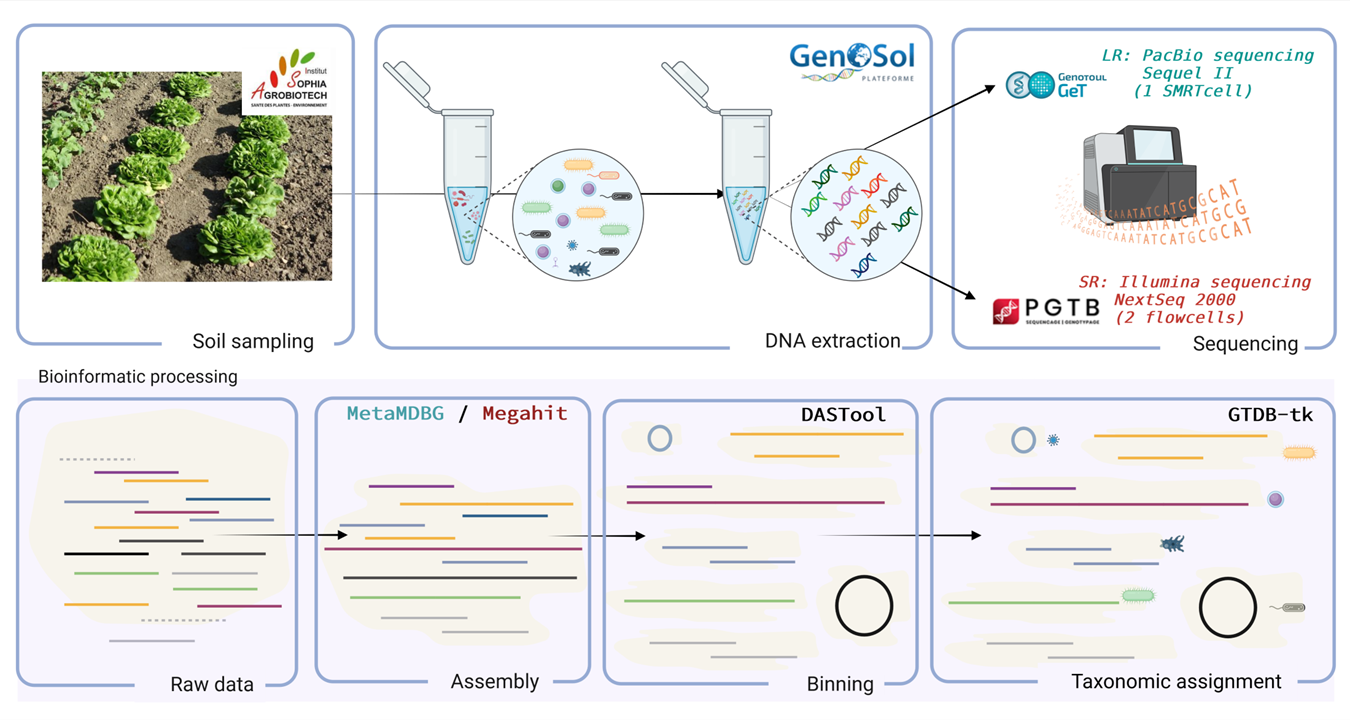
Caption
Metagenomic analysis protocol of soil for the reconstruction of genomes of microbial species present in a sample.
- Collection of a soil sample from a cultivated plot in southern France by the teams of the Institut Sophia Agrobiotech.
- Extraction of DNA molecules by the Genosol platform.
- DNA sequencing:
- Long-read sequencing: PacBio HiFi Sequel II (1 flowcell) by the GetPlage platform at INRAe Toulouse.
- Short-read sequencing: Illumina NextSeq 2000 2x150 bp by the PGTB platform at INRAe Bordeaux.
- Bioinformatic analysis:
- Quality control and analysis of raw data to eliminate erroneous or low-quality sequences.
- Genome reconstruction: High-quality reads are then assembled to reconstruct the genomes of the microorganisms present in the sample.
- Binning of assembled DNA fragments to group them by genomes. This allows the association of sequences according to the organism of origin.
- Taxonomic analysis to identify the species present in the sample. This step uses reference databases and methods of homology searching and phylogenetic placement to classify microorganisms and understand the microbial diversity of the studied soil.

